Proposition de thèse financée par l’ERC
Study of the relationship between selection and genetic architecture of traits in the common lizard
Offer Description
Summary
We offer a 3-years position, working as a PhD candidate to study the link between selection, local adaptation and the genetic architecture of complex traits on the common lizard (Zootoca vivipara). The candidate will pass their PhD at École Pratique des Hautes Études (PSL University), joining Pierre de Villemereuil’s team, at the Institute for Systematics, Evolution, Biodiversity (ISYEB), located within the Muséum National d’Histoire Naturelle (MNHN) in Paris. This position is funded for 3-years as part of the EvoGenArch ERC Starting Grant (n°101114976). The PhD is planned to start in October 2025, although starting date can be discussed.
Keywords evolutionary quantitative genetics, population genomics, evolutionary ecology, genetic architecture, adaptation, Zootoca vivipara
Scientific context
Most traits of ecological and evolutionary relevance are complex traits, i.e. influenced by a large number of loci (named Quantitative Trait Loci, or QTL) in the genomes (Lynch & Walsh, 1998; Walsh & Lynch, 2018). The features of the QTL are often referred to as the genetic architecture of the trait. One of the most important collective feature to characterise such architecture is the distribution of the effect sizes (i.e. the phenotypic impact of replacing one allele for the other) of QTL. While theoretical debates surrounding the typical distribution of such effect sizes can be dated back to the origins of neo-darwinism (Orr, 2005), the recent theoretical literature tackling this subject has shown that the evolution of such distribution is highly dependent on precise assumptions regarding neutral and selective forces, as well as the genetic variation to evolve from (Yeaman, 2015; Dittmar et al., 2016; Höllinger et al., 2019). Empirical data to support this link between the features of evolution and the resulting shape of the distribution of effect sizes for various traits is however lacking. Beyond the theoretical controversy highlighted above, another reason for this lack of empirical data is the problem that current methods allowing for the study of such distribution of effect sizes make the same core assumption: namely, that all QTL are a subset of the genetic markers (generally SNPs) available in the genome, although QTL can in theory be any kind of variant. In the context of the EvoGenArch project, we are developing a new statistical method aiming at solving this issue, using recombination information through the information contained in Ancestral Recombination Graphs (ARGs).
PhD project description
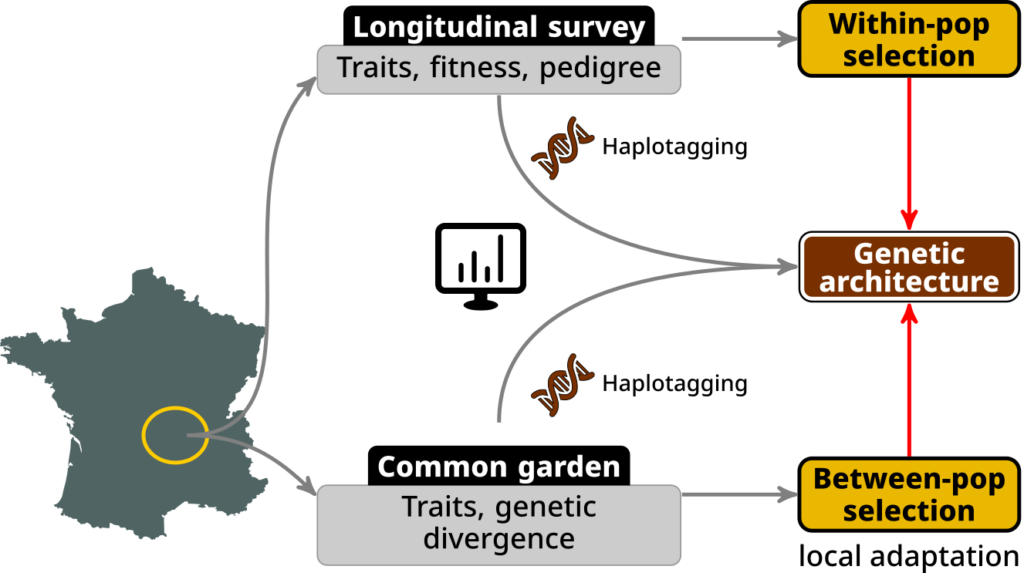
The purpose of this PhD is to study the link between selection and the distribution of effect sizes, using the method developed within the context of the EvoGenArch project, large datasets on a wild and a common garden populations of the common lizard (Z. vivipara), each with a thousand individuals whole-genome sequenced using a linked-read approach named haplotagging.

The common lizard is a model species in evolution and ecology, intensively studied in various context such as ecophysiology (Dupoué et al., 2020), behavioural ecology (Chabaud et al., 2023), evolutionary ecology (Bodineau et al., 2024) and evolutionary genetics (San-Jose et al., 2023). This species can be studied in wild populations (Rutschmann et al., 2021; Dupoué et al., 2022), as well as in captive enclosed populations within experimental setups (Bestion et al., 2015, 2023). The project will first use a >35 years-old longitudinal data on a wild population atop Mt Lozère in France, with pedigree information since 2000. Second, the project will use data from a common garden experiment, started in 2022, using offspring from females caught in 14 populations of the French Massif Central. This population is also pedigreed.
In each of these settings, c.a. thousand individuals will be whole-genome sequenced using a linked-read approach named haplotagging (Meier et al., 2021). Such approach will allow for a precise and local reconstruction of the haplotypes of individuals and thus provide excellent information for the inference of ARGs on which the statistical method to infer the distribution of the effect sizes of QTL is based on.
The PhD candidate will use all of these data to :
- Infer the “within-population” selection regime (e.g. directional v. stabilising, fluctuating or not) of the different traits in the wild population,
- Analyse the whole-genome haplotagging data for both the wild and enclosure populations, including performing kinship inferences to marginally correct both pedigrees if required and be used in the next step,
- Infer the “between-population” (i.e. local adaptation) acting on the traits based on the common garden experiment, notably using kinship inferences from the previous step,
- Use the method developed within the EvoGenArch project (by a hired post-doc) to analyse the genetic architecture (i.e. distribution of effect sizes) of the measured traits,
- Relate the diversity of inferred genetic architectures to the within- and between-population selection acting on the traits.
The PhD candidate will have freedom to refine each of the cursory steps detailed above. Beside, while the above steps are well circumscribed, the large amount of data available and generated during the project will provide the candidate a large amount of room to provide new ideas and scientific prospects of analysis. They will participate to the molecular and bio-informatics parts of the haplotagging protocol, under the supervision of the team technicians and engineers. They will also participate to the two-month field work in the French Massif Central, each summer.
Hosting lab & team
Pierre de Villemereuil’s research focuses on the genetics of adaptation, trying to understand how the levels of the genotype, phenotype and environment interacts to drive evolution in wild populations, notably in response to anthropic pressures (de Villemereuil, Gaggiotti, et al., 2016; de Villemereuil, Schielzeth, et al., 2016; de Villemereuil et al., 2018, 2019, 2020, 2022; Bonnet et al., 2022; Salloum et al., 2022), using a combination of evolutionary ecology, quantitative genetics, population genomics and statistical modelling. The Institute for Systematics, Evolution, Biodiversity (ISYEB), located in the beautiful Jardin des Plantes of the Muséum National d’Histoire Naturelle in Paris,is one of the largest labs studying evolution in the city. ISYEB hosts worldwide leading science in evolution, systematics, phylogeny, genomics and ecology.
Requirements
- A Master in evolutionary genetics or evolutionary ecology is required.
- Background in population genetics and/or quantitative genetics is required.
- Experience and/or training in working with genomic data will be strongly appreciated.
- Experience/skills in using a shell in a Unix environment will be appreciated.
- Background in quantitative genetics and/or evolutionary ecology will be appreciated.
- Motivation for the scientific study of evolution and genetics.
- Motivation for participating in the field work, working with animals in the context of a scientific protocol and tend to their needs.
- Ability to work as part of a team within a scientific project.
Selection process
Please send, before May 13th, (1) a CV/résumé; (2) a short (max. 2 pages) letter of application highlighting your fit to the profile and your motivation for the position; and (3) a letter of recommendation of past supervisor or teacher to the address pierre.de-villemereuil@mnhn.fr. Selected candidates will be auditioned by a committee of two to three collaborators on the project, including the PhD supervisor.
References
- Bestion, E., San-Jose, L. M., Di Gesu, L., Richard, M., Sinervo, B., Côte, J., Calvez, O., Guillaume, O., & Cote, J. (2023). Plastic responses to warmer climates: A semi-natural experiment on lizard populations. Evolution, 77(7), 1634–1646. https://doi.org/10.1093/evolut/qpad070
- Bestion, E., Teyssier, A., Richard, M., Clobert, J., & Cote, J. (2015). Live fast, die young: Experimental evidence of population extinction risk due to climate change. PLOS Biology, 13(10), e1002281. https://doi.org/10.1371/journal.pbio.1002281
- Bodineau, T., de Villemereuil, P., Agostini, S., Decencière, B., Le Galliard, J.-F., & Meylan, S. (2024). Breeding phenology drives variation in reproductive output, reproductive costs, and offspring fitness in a viviparous ectotherm. Journal of Evolutionary Biology, 37(9), 1023–1034. https://doi.org/10.1093/jeb/voae086
- Bonnet, T., Morrissey, M. B., de Villemereuil, P., Alberts, S. C., Arcese, P., Bailey, L. D., Boutin, S., Brekke, P., Brent, L. J. N., Camenisch, G., Charmantier, A., Clutton-Brock, T. H., Cockburn, A., Coltman, D. W., Courtiol, A., Davidian, E., Evans, S. R., Ewen, J. G., Festa-Bianchet, M., … Kruuk, L. E. B. (2022). Genetic variance in fitness indicates rapid contemporary adaptive evolution in wild animals. Science, 376(6596), 1012–1016. https://doi.org/10.1126/science.abk0853
- Chabaud, C., Lourdais, O., Decencière, B., & Le Galliard, J.-F. (2023). Behavioural response to predation risks depends on experimental change in dehydration state in a lizard. Behavioral Ecology and Sociobiology, 77(7), 90. https://doi.org/10.1007/s00265-023-03362-2
- de Villemereuil, P., Charmantier, A., Arlt, D., Bize, P., Brekke, P., Brouwer, L., Cockburn, A., Côté, S. D., Dobson, F. S., Evans, S. R., Festa-Bianchet, M., Gamelon, M., Hamel, S., Hegelbach, J., Jerstad, K., Kempenaers, B., Kruuk, L. E. B., Kumpula, J., Kvalnes, T., … Chevin, L.-M. (2020). Fluctuating optimum and temporally variable selection on breeding date in birds and mammals. Proceedings of the National Academy of Sciences, 117(50), 31969–31978. https://doi.org/10.1073/pnas.2009003117
- de Villemereuil, P., Gaggiotti, O. E., & Goudet, J. (2022). Common garden experiments to study local adaptation need to account for population structure. Journal of Ecology, 110(5), 1005–1009. https://doi.org/10.1111/1365-2745.13528
- de Villemereuil, P., Gaggiotti, O. E., Mouterde, M., & Till-Bottraud, I. (2016). Common garden experiments in the genomic era: New perspectives and opportunities. Heredity, 116(3), 249–254. https://doi.org/10.1038/hdy.2015.93
- de Villemereuil, P., Mouterde, M., Gaggiotti, O. E., & Till-Bottraud, I. (2018). Patterns of phenotypic plasticity and local adaptation in the wide elevation range of the alpine plant Arabis alpina. Journal of Ecology, 106(5), 1952–1971. https://doi.org/10.1111/1365-2745.12955
- de Villemereuil, P., Rutschmann, A., Lee, K. D., Ewen, J. G., Brekke, P., & Santure, A. W. (2019). Little adaptive potential in a threatened passerine bird. Current Biology, 29(5), 889-894.e3. https://doi.org/10.1016/j.cub.2019.01.072
- de Villemereuil, P., Schielzeth, H., Nakagawa, S., & Morrissey, M. B. (2016). General methods for evolutionary quantitative genetic inference from generalised mixed models. Genetics, 204(3), 1281–1294. https://doi.org/10.1534/genetics.115.186536
- Dittmar, E. L., Oakley, C. G., Conner, J. K., Gould, B. A., & Schemske, D. W. (2016). Factors influencing the effect size distribution of adaptive substitutions. Proceedings of the Royal Society B: Biological Sciences, 283(1828), 20153065. https://doi.org/10.1098/rspb.2015.3065
- Dupoué, A., Blaimont, P., Angelier, F., Ribout, C., Rozen-Rechels, D., Richard, M., Miles, D., de Villemereuil, P., Rutschmann, A., Badiane, A., Aubret, F., Lourdais, O., Meylan, S., Cote, J., Clobert, J., & Le Galliard, J.-F. (2022). Lizards from warm and declining populations are born with extremely short telomeres. Proceedings of the National Academy of Sciences, 119(33), e2201371119. https://doi.org/10.1073/pnas.2201371119
- Dupoué, A., Blaimont, P., Rozen-Rechels, D., Richard, M., Meylan, S., Clobert, J., Miles, D. B., Martin, R., Decencière, B., Agostini, S., & Le Galliard, J.-F. (2020). Water availability and temperature induce changes in oxidative status during pregnancy in a viviparous lizard. Functional Ecology, 34(2), 475–485. https://doi.org/10.1111/1365-2435.13481
- Höllinger, I., Pennings, P. S., & Hermisson, J. (2019). Polygenic adaptation: From sweeps to subtle frequency shifts. PLOS Genetics, 15(3), e1008035. https://doi.org/10.1371/journal.pgen.1008035
- Lynch, M., & Walsh, B. (1998). Genetics and analysis of quantitative traits. Sinauer Associates.
- Meier, J. I., Salazar, P. A., Kučka, M., Davies, R. W., Dréau, A., Aldás, I., Power, O. B., Nadeau, N. J., Bridle, J. R., Rolian, C., Barton, N. H., McMillan, W. O., Jiggins, C. D., & Chan, Y. F. (2021). Haplotype tagging reveals parallel formation of hybrid races in two butterfly species. Proceedings of the National Academy of Sciences, 118(25). https://doi.org/10.1073/pnas.2015005118
- Orr, H. A. (2005). The genetic theory of adaptation: A brief history. Nature Reviews Genetics, 6(2), 119–127. https://doi.org/10.1038/nrg1523
- Rutschmann, A., Dupoué, A., Miles, D. B., Megía-Palma, R., Lauden, C., Richard, M., Badiane, A., Rozen-Rechels, D., Brevet, M., Blaimont, P., Meylan, S., Clobert, J., & Le Galliard, J.-F. (2021). Intense nocturnal warming alters growth strategies, colouration and parasite load in a diurnal lizard. Journal of Animal Ecology, 90(8), 1864–1877. https://doi.org/10.1111/1365-2656.13502
- Salloum, P. M., Santure, A. W., Lavery, S. D., & de Villemereuil, P. (2022). Finding the adaptive needles in a population-structured haystack: A case study in a New Zealand mollusc. Journal of Animal Ecology, 91(6), 1209–1221. https://doi.org/10.1111/1365-2656.13692
- San-Jose, L. M., Bestion, E., Pellerin, F., Richard, M., Di Gesu, L., Salmona, J., Winandy, L., Legrand, D., Bonneaud, C., Guillaume, O., Calvez, O., Elmer, K. R., Yurchenko, A. A., Recknagel, H., Clobert, J., & Cote, J. (2023). Investigating the genetic basis of vertebrate dispersal combining RNA-seq, RAD-seq and quantitative genetics. Molecular Ecology, n/a(n/a). https://doi.org/10.1111/mec.16916
- Walsh, B., & Lynch, M. (2018). Evolution and selection of quantitative traits. Oxford University Press.
- Yeaman, S. (2015). Local adaptation by alleles of small effect. The American Naturalist, 186(S1), S74–S89. https://doi.org/10.1086/682405
This action has received funding from the European Union’s Horizon research and innovation program & European Research Council under grant agreement n°101114976 – EvoGenArch.

