Population structure in common gardens! (and a Ph.D. project?)
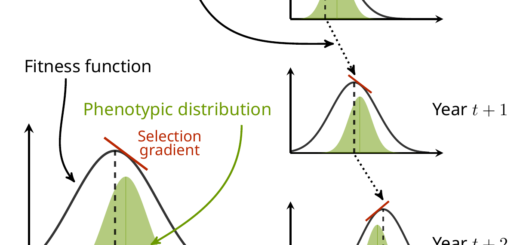
Common garden experiments are useful to study the genetics of local adaptation, because they allow to compare individuals from different populations of origin within the same environmental conditions, thus overcoming the confounding effect of phenotypic plasticity in the wild.
A common framework to test for the presence of local adaptation using a common garden experiment is to compare the QST (a genetic differentiation index based on variance decomposition from the observed phenotypic values) to the FST (a genetic differentiation index based on neutral allelic frequencies, e.g. from molecular data). Since its inception in the 90s, the QST-FST comparison framework has become a major tool to test and study local adaptation.
With Oscar Gaggiotti and Jérôme Goudet, we explored a bit the consequences of population structure on the inference of QST and its comparison to FST in a mini-review for Journal of Ecology. We explain the assumptions behind such computation and show that population structure can have a strong impact on the error rate for QST-FST comparison. We detail two existing solution aiming at, at least partly, overcoming the issue: one from Ovaskainen et al. (2011) and the other from Josephs et al. (2017).
If you are a student looking for a Ph.D. project, we (Jérôme, Oscar and myself) would like to push the study of the impact of population structure on both the estimation of QST and FST, and develop further methodologies that would be more robust to such impact, and believe this would be a very nice Ph.D. project to be supervised by the three of us. Candidates should ideally have an interest for quantitative genetics, population genetics and/or statistical inference. Given our different locations (me in Paris, Jérôme in Lausanne and Oscar in St-Andrews), the project would also allow the student to discover different (but equally dynamic) academic places in different (but equally nice) countries!