Quantitative genetics of reaction norms: an onion partitionning
So… this project dates back from… 2018 ?! When I started my post-doc with Luis-Miguel Chevin, we discussed variance partitioning in the context of phenotypic plasticity, especially around some limitations we saw in a meta-analysis paper published in 2014. We wanted to design a variance partition that could account for the contribution of the “shape” of reaction norms to the total phenotypic variance. It took us 7 years to publish it and it took me almost a year to write up something about it on this site…

Why partition variance of phenotypic plasticity?
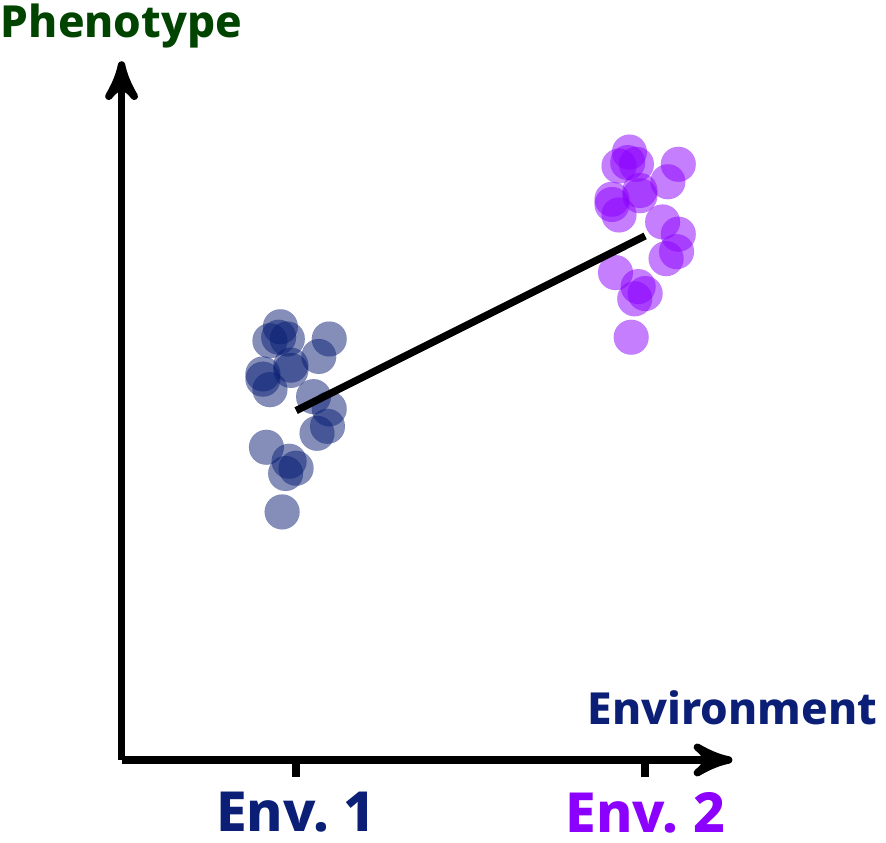
Phenotypic traits are often plastic, meaning that how they are expressed depend on the kind of environment they are expressed in. A way that we can study such phenotypic plasticity is by using reaction norms, i.e. the prediction of the average phenotypic value a trait will take in a given environment, for a given genotype. Mathematically, we can define such reaction norm as a function that takes environmental and genotypic values and outputs a phenotypic value, something like the black line in the following graph on the left.
However, the shape is not necessarily just a straight line (although this is a widespread simplifying assumption): in theory, it could take any kind of shape. And so, the question we tried to answer was: was if we wanted to measure the phenotypic variance produced by that black line (and variation of such black line among different genotypes) in the most general case, and would we do it?
It turned out, the question is not that easy to answer, and there are many questions, concepts and obstacles nested within this yet very easily formulated objective. When first tackling the subject, we (Luis-Miguel and I) did not expect to not just go down the rabbit hole, but dig a whole gallery large enough to host a city-full of rabbit families!
That’s basically the main reason why it took us that long to finish this project and publish the paper (that, and also, it was a side project for the both of us, which didn’t help). For each solution we found, we always discovered new layers of complexity we wanted to explore… and had to take some time to argue over them again and again until we finally agreed (or agreed to disagree, sometimes)! This dialectic process was, in the end, very fruitful, thought-provoking and full of exciting revelations and discoveries.
How does it work?
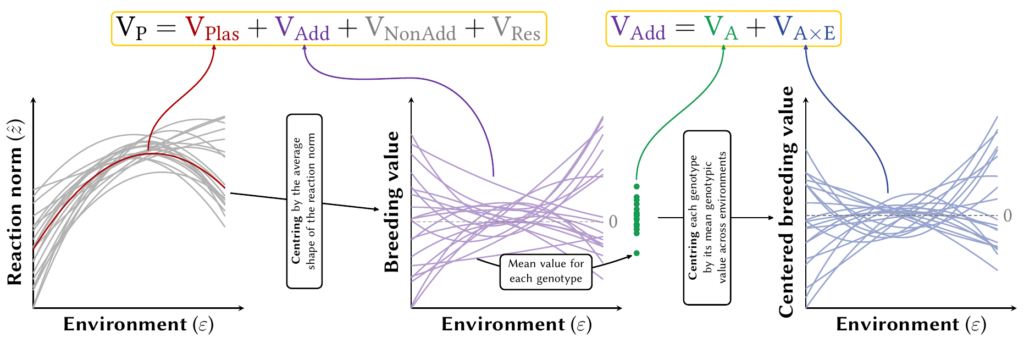
The first of our framework is to decompose the total phenotypic variance into 3 components:
- The variance arising from the average shape of the reaction norm across all the different genotypes, which we called VPlas;
- The genetic variance arising from the variation among genotypes beyond that average shape, which we called VGen;
- A residual variance, for the variation in the phenotypic trait that is not predicted by the reaction norm, which we called VRes.
Because we are evolutionist, and thus interested in response to selection, we focused on the part of the genetic variance VGen that is heritable (additive), which we called VAdd. And we further decomposed (I was taking about layers…) that total heritable variance VAdd into the “environment-blind” heritable variance (that would be computed if we ignored phenotypic plasticity), VA, on the one hand; and the heritable variance arising from phenotypic plasticity, which we called VAxE. By dividing those quantities by the total variance, we can compute the proportion of the variance due to VPlas (P²RN) and the corresponding heritabilities for the heritable components (h²RN, h² and h²I).
But this not where the layers stop: we can further attribute how much these variances depend on the shape or parameters of the reaction norms. This true both of VPlas and of the heritable components (VAdd, VA, VAxE).
Regarding the variance due to the average shape of the reaction norm (VPlas), we can decompose it into the variance arising from the average slope, or from the average curvature, of the reaction norm based on some assumptions (e.g. normality). We called that one the π-decomposition. The heritable components (VAdd, VA, VAxE) are, by definition, additive (for reasons I can’t get into right now). This means that their computation is basically a huge sum of products (a linear combination) which depends on the environment and the parameters of the reaction norm. Each of the products can be related back to one, or a pair of, parameter(s) of the reaction norm. This means that we can assess how much of each of these heritable components depend on genetic variation in the reaction norm parameters. We called those the γ-decomposition for VAdd and the ι-decomposition for VAxE.
What’s the point?
With this framework, we provide a few things:
- We believe it clears up a few things on the relationship between reaction norms and phenotypic variance. Some of it was already known but not shown exactly like we did. Some of it we believe to be fairly new, at least to such a general extent.
- It provides a way to standardise the contribution of the shape of reaction norms to the phenotypic variance, which (remember) was our first and main goal.
- It provides an extremely general and theoretically grounded framework to perform the quantitative genetic study of reaction norms, and especially, explore where the heritable adaptive potential lie within them (in the environmental-blind evaluation of the trait? In plasticity? Which parameter of the reaction norm contribute the more to it?).
- It is designed so that all of the above can be applied in a maximally general way, accounting for all of the different ways we use to handle reaction norms (using linear or non-linear curves, or a discretised approach named “character-state”).
To maximise adoption, we provide all of the framework as a nice set of functions put together in the Reacnorm R package (see also the Github repository if needed).
Where is this published?
As both Luis-Miguel and I have permanent positions (and no longer have to have as much shiny journal names in our CV as possible), we decided to support the Peer Community In initiative with this publication. This initiative is meant to circumvent the highly dysfunctional economy of scientific publication by promoting a process of peer recommendation based on pre-prints, optionally followed by publication in the online Peer Community Journal where our paper is now published.
I have to say the process of publication was extremely satisfying, with a lot of the frictions relieved, both during the review and post-review processes. The reviewing was just as strict and challenging as for any serious journal (the review process is transparent and available online if you want to check). We had a lot of control on the editing of the final version of the paper, with cordial dialogue with the editorial team, which was a very refreshing change from many other venues for publication! 5/5 ⭐️⭐️⭐️⭐️⭐️, recommend!
It does mean that our paper might escape some of its current readership though (not used to monitor such publication venue), so please help us by making our paper known around you if you like what’s in it! 🙏